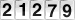Join NPTEL course on Host - Pathogen Interaction (Immunology)
Innate immune sensing of influenza A viral RNA through IFI16
Cytosolic DNA sensor IFI16 is essential for the activation of programmed cell death pathways in influenza virus infected cells. IFI16 functions as an RNA sensor for the influenza A virus by interacting with genomic RNA.
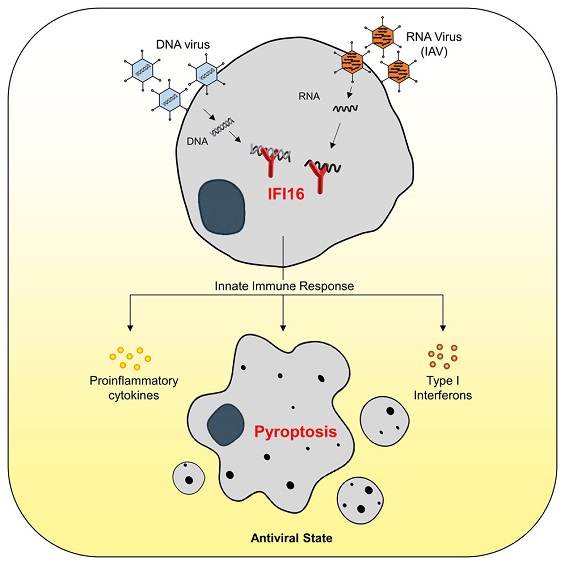
NIX-mediated mitophagy regulate metabolic reprogramming during mycobacterial infection
NIX knockdown abrogates upregulation of glycolysis, mitophagy, and secretion of pro-inflammatory cytokines. Mycobacterial infection-induced immunometabolic changes are executed via NIX mediated mitophagy and are essential for macrophage differentiation and resolution of infection.
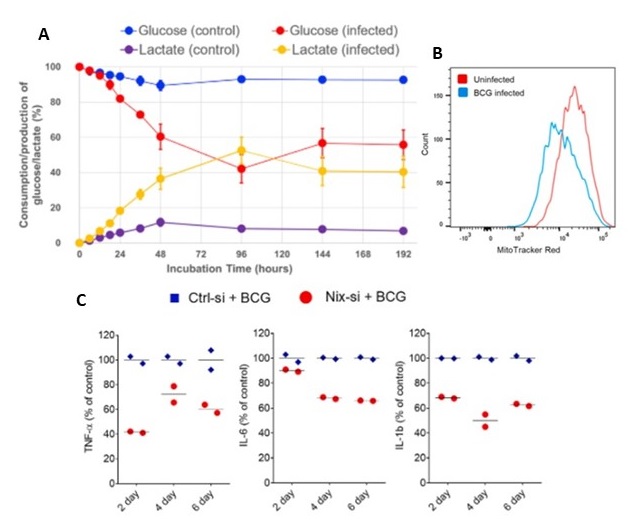
Comparative transcriptome analysis of SARS-CoV, MERS-CoV, and SARS-CoV-2
SARS-CoV-2 mostly alters the host pathways similar to SARS-CoV and MERS-CoV. COVID-19 patients show altered ribosomes and mitochondrial pathways. The drugs to be targeting these pathways for the treatment using co-gene expression and drug repurposing analysis identified potential drug candidates against COVID-19.
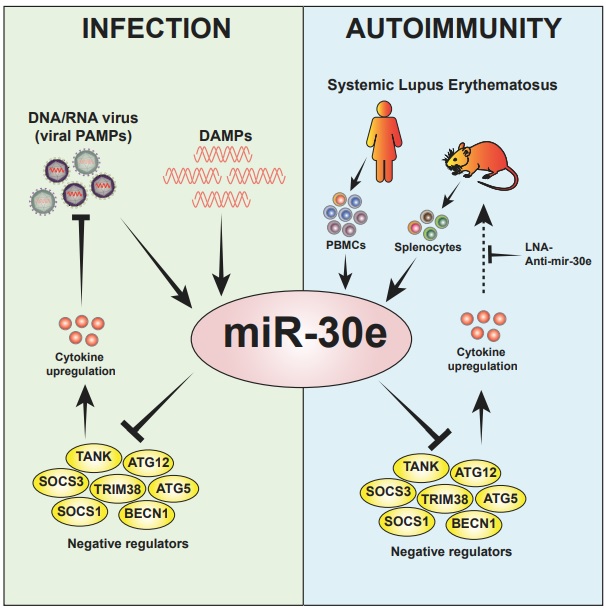
Integrated Role of miRNA-30e-5p in Regulation of the Innate Immune Response
miR-30e targets multiple negative regulators of innate immune signaling and its sequestering in SLE patients reduces type-I interferon and pro-inflammatory cytokines.
Particulate matter enhances RNA virus infection
Air pollution skews innate immunity during Influenza virus infection by suppressing anti-viral innate immunity
H5N1 downregulates miR-324-5p expression to promote viral evasion
(A) Multiple-sequence alignment of the target site of miR-324-5p in the PB1 gene across different strains of influenza A virus. (B) Model showing that miR-324-5p binds to the 3' UTR of host CUEDC2 and viral PB1 and subsequently inhibits the replication of H5N1.
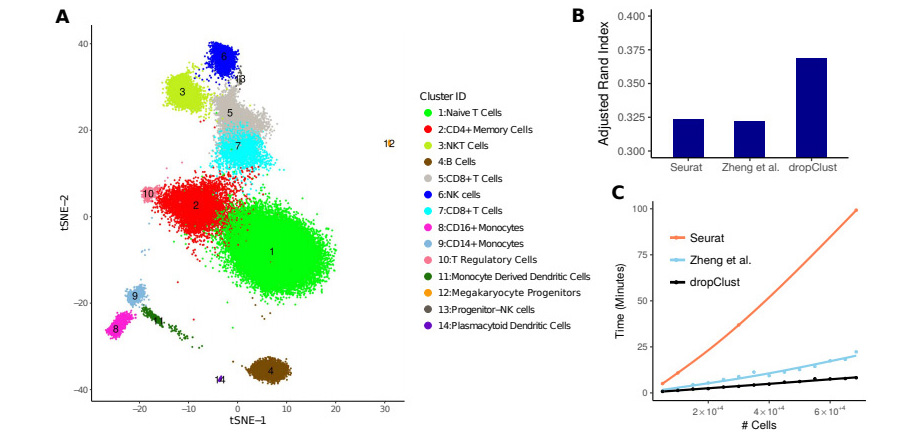
Clustering of ~68k PBMC data using dropClust
(A) dropClust based visualization of the transcriptomes. (B) Bars show the ARI indexes obtained by comparing clustering outcomes with cell-type annotations. (C) Trend of increase in execution time for different clustering methods.
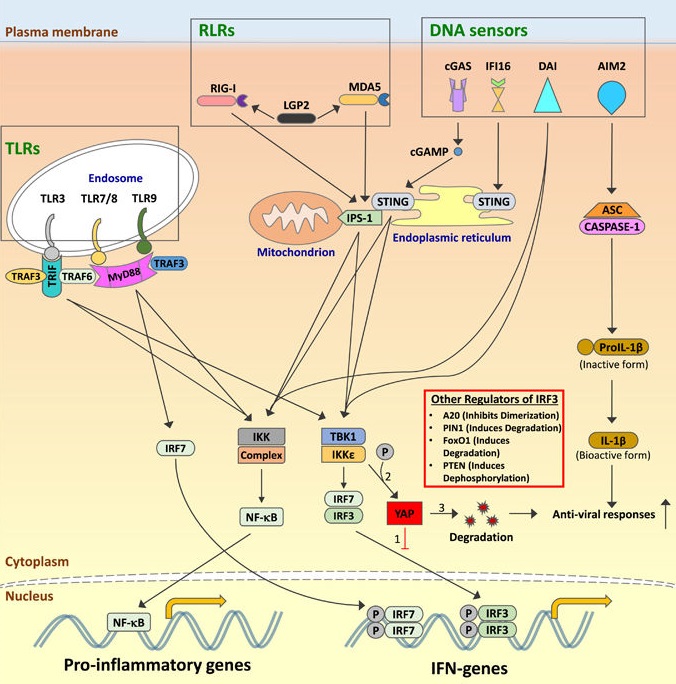
Balancing anti-viral innate immunity and immune homeostasis
Schematic representation of the regulation of the anti-viral signaling pathway, after sensing by viral products by their cognate pattern recognition receptors (PRRs).
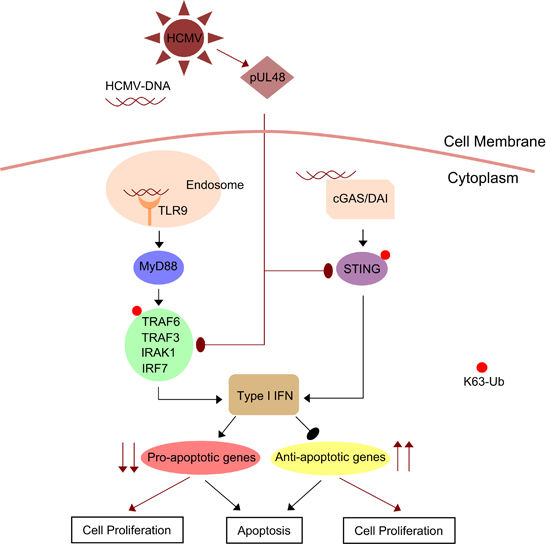
Induction of oncogenesis by HCMV deubiqutinase
HCMV-DUB facilitates deubiquitination of TRAF6, TRAF3, IRAK1, IRF7 and STING to inhibit I-IFN synthesis, which in turn inhibits the expression of several pro-apoptotic genes and induces the expression of anti-apoptotic genes.
Bispecific targeting of the 3′UTR of RIG-I and PB1 by miR-485.
A) Model showing the targeting of RIG-I by miR-485 and the subsequent effects on antiviral responses and the replication of NDV and H5N1 when present at low viral titers. (B) Model showing the targeting of H5N1 PB1 by miR-485 and the subsequent effects on antiviral responses and H5N1 replication at high viral titers.
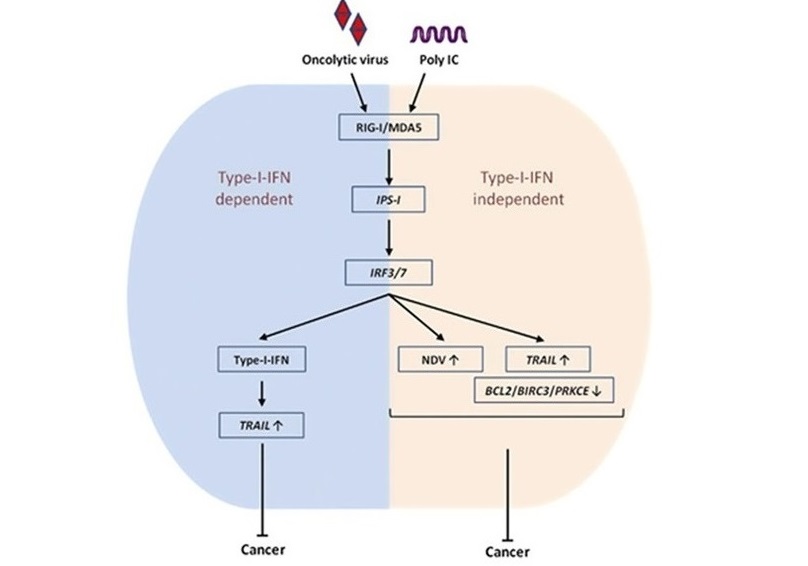
Regulatory role of IPS1 in cancer ablation
NDV infection and polyIC transfection induce type I IFN-dependent and -independent anticancer activity via the IPS-1, IRF3 and IRF7 axis. The mechanism of the type I IFN relies on high NDV replication, upregulation of TRAIL or downregulation of the BCL2, BIRC3 and PRKCE genes